ERGO: Efficient Recurrent Graph Optimized Emitter Density Estimation in Single Molecule Localization Microscopy
 ERGO
ERGO
ERGO: Efficient Recurrent Graph Optimized Emitter Density Estimation in Single Molecule Localization Microscopy
Abstract
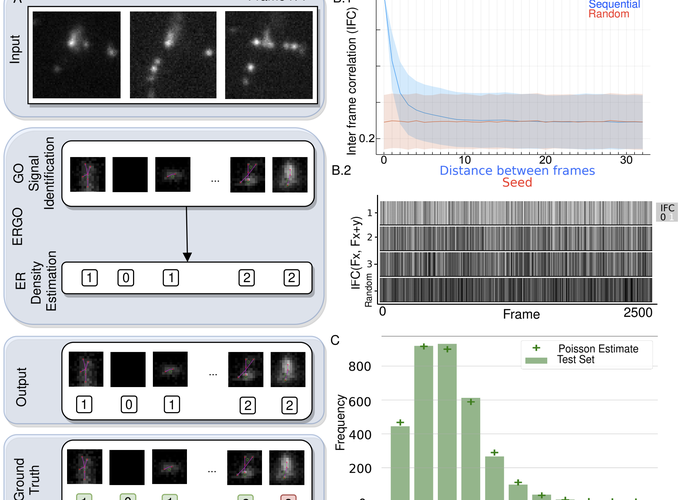
Single molecule localization microscopy (SMLM) allows unprecedented insight into the three-dimensional organization of proteins at the nanometer scale. The combination of minimal invasive cell imaging with high resolution positions SMLM at the forefront of scientific discovery in cancer, infectious, and degenerative diseases. By stochastic temporal and spatial separation of light emissions from fluorescent labelled proteins, SMLM is capable of nanometer scale reconstruction of cellular structures. Precise localization of proteins in 3D astigmatic SMLM is dependent on parameter sensitive preprocessing steps to select regions of interest. With SMLM acquisition highly variable over time, it is non-trivial to find an optimal static parameter configuration. The high emitter density required for reconstruction of complex protein structures can compromise accuracy and introduce artifacts. To address these problems, we introduce two modular auto-tuning pre-processing methods: adaptive signal detection and learned recurrent signal density estimation that can leverage the information stored in the sequence of frames that compose the SMLM acquisition process. We show empirically that our contributions improve accuracy, precision and recall with respect to the state of the art. Both modules auto-tune their hyper-parameters to reduce the parameter space for practitioners, improve robustness and reproducibility, and are validated on a reference in silico dataset. Adaptive signal detection and density prediction can offer a practitioner, in addition to informed localization, a tool to tune acquisition parameters ensuring improved reconstruction of the underlying protein complex. We illustrate the challenges faced by practitioners in applying SMLM algorithms on real world data markedly different from the data used in development and show how ERGO can be run on new datasets without retraining while motivating the need for robust transfer learning in SMLM.